Aberrant DNA methylation, a type of epigenetic modification, is a common hallmark of tumor biology. DNA methylation refers to the addition of a methyl group to the DNA molecule, which can alter gene expression without changing the underlying DNA sequence. This process plays a critical role in normal development and cellular differentiation of cells. However, in cancer cells, aberrant DNA methylation can result in the silencing of tumor suppressor genes and the activation of oncogenes, contributing to tumor formation and progression.
DNA methylation is a chemical modification of the DNA structure caused by enzymes. A methyl (CH3) group is covalently bonded to the cytosine base’s 5-carbon. This process is mediated by one or more of the DNA methyltransferase enzymes. S-adenosyl methionine (SAM) provides the methyl group, which is converted to S-adenosyl homocysteine (SAH) during the process. In a folate- and cobalamin-dependent pathway, this is recycled back to SAM. In vertebrates, biologic methylation occurs only on cytosine bases that are linked directly to guanine by the phosphodiesterase link, forming a CpG dinucleotide pair.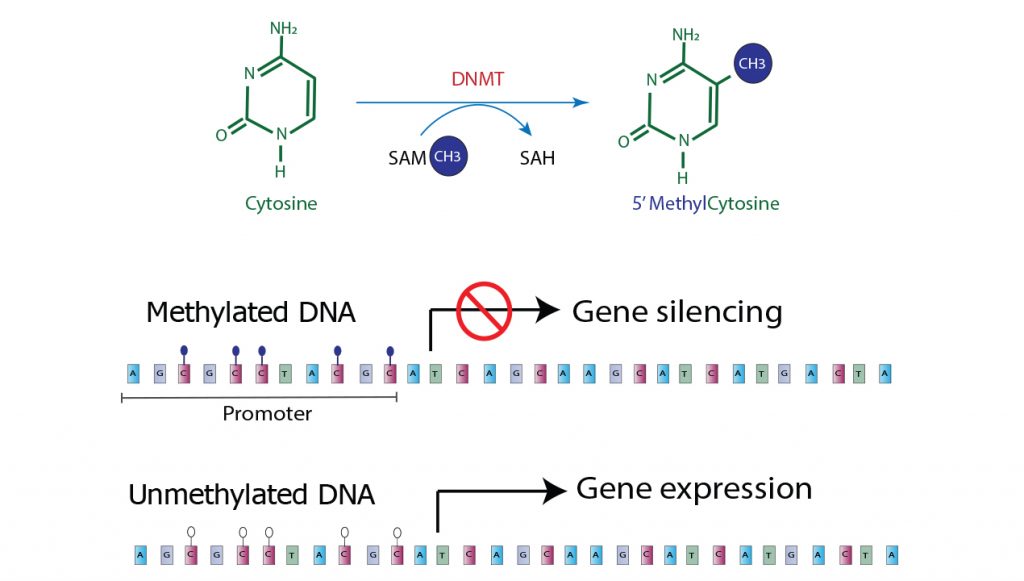
DNA methylation is now recognized as an essential and normal component of mammalian embryogenesis. Targeted mutations of DNA methyltransferase genes in mice result in early embryonic lethality by knocking out or drastically reducing enzyme production and methylation activity.
It is possible for a fetus to survive with a moderately reduced level of methyltransferase because it causes less severe disruption of methylation patterns. The uncommon ICF syndrome (immunodeficiency, centromeric heterochromatin instability and facial anomalies) is caused by mutations in the DNMT3B gene, which codes for a methyltransferase thought to be specifically responsible for specific methylation of centromeric satellite repeat sequences.
Once a new methylation pattern is established throughout the embryonic genome, it remains constant and is maintained throughout life. This function allows the cell to regulate its own activity by turning off the expression of specific genes when they are not needed.
Cancer cells are distinguished by the formation of an abnormal DNA methylation pattern. These DNA methylation abnormalities are thought to play a role in cancer development. Identifying the specific DNA methylation abnormalities that contribute to tumor formation and progression can provide important insights for developing effective therapeutic strategies. Therefore, understanding the role of aberrant DNA methylation in cancer biology is crucial for improving cancer diagnosis, prognosis and treatment.
Cancer cells’ DNA has been shown to have altered methylation patterns. There are two patterns that have been observed: large areas of global hypomethylation along the genome and localized areas of hypermethylation at specific sites, the CpG islands, within gene promoter regions.
According to basic genetic models of cancer, when proto-oncogenes are amplified or tumor suppressor genes are silenced, it disrupts the natural balance that controls cell growth and proliferation, leading to the development of cancer. In theory, reducing DNA methylation levels could reverse the transcriptional silencing and activate proto-oncogenes that were previously inactive, triggering the cellular proliferation that drives cancer progression. Therefore, targeting DNA methylation could be a promising approach for treating cancer by altering the gene expression patterns and restoring the normal balance of cell growth and proliferation.
Alternatively, if certain regions of DNA, such as the promoter regions of tumor suppressor genes, become abnormally methylated, it can silence the genes and prevent them from inhibiting cell proliferation. This occurs because the methylation can block the gene’s transcription and reduce its expression, allowing the uncontrolled growth and division of cells that contribute to the development of cancer. Therefore, understanding the role of DNA methylation in regulating gene expression again is crucial for developing effective cancer therapies that target the underlying molecular mechanisms.
Abnormal methylation patterns can also have an indirect effect on gene activity by disrupting the transcription-translation process and increasing the likelihood of a mutational event occurring, resulting in the production of a dysfunctional protein product. Methylated cytosine is more prone to spontaneous deamination and the formation of thymine. If this happens on a tumor suppressor gene, a point mutation develops and cell proliferation control is lost.
If it is consistently observed that DNA methylation of gene promoter regions is an early event in the development of cancer, with specific gene combinations for different types of cancer, it could provide a tool for diagnosing premalignant lesions. This would enable more accurate screening and surveillance of patients at high risk for developing cancer on a molecular basis. It would also provide a basis for definitive treatment of patients who have molecular but not yet all the typical pathologic or microscopic features associated with cancer. Removing the affected tissue through surgical procedures before it progresses to an invasive and metastatic stage could prevent subsequent cancer in such patients. This could be done through minimally invasive procedures that are associated with fewer complications. Therefore, understanding the role of DNA methylation in cancer development has the potential to transform cancer diagnosis and treatment by enabling early detection and intervention.
References:
- Lakshminarasimhan R, Liang G. The Role of DNA Methylation in Cancer. Adv Exp Med Biol. 2016;945:151-172. doi:10.1007/978-3-319-43624-1_7
- Wajed SA, Laird PW, DeMeester TR. DNA methylation: an alternative pathway to cancer. Ann Surg. 2001;234(1):10-20. doi:10.1097/00000658-200107000-00003

MDForLives is a global healthcare intelligence platform where real-world perspectives are transformed into validated insights. We bring together diverse healthcare experiences to discover, share, and shape the future of healthcare through data-backed understanding.