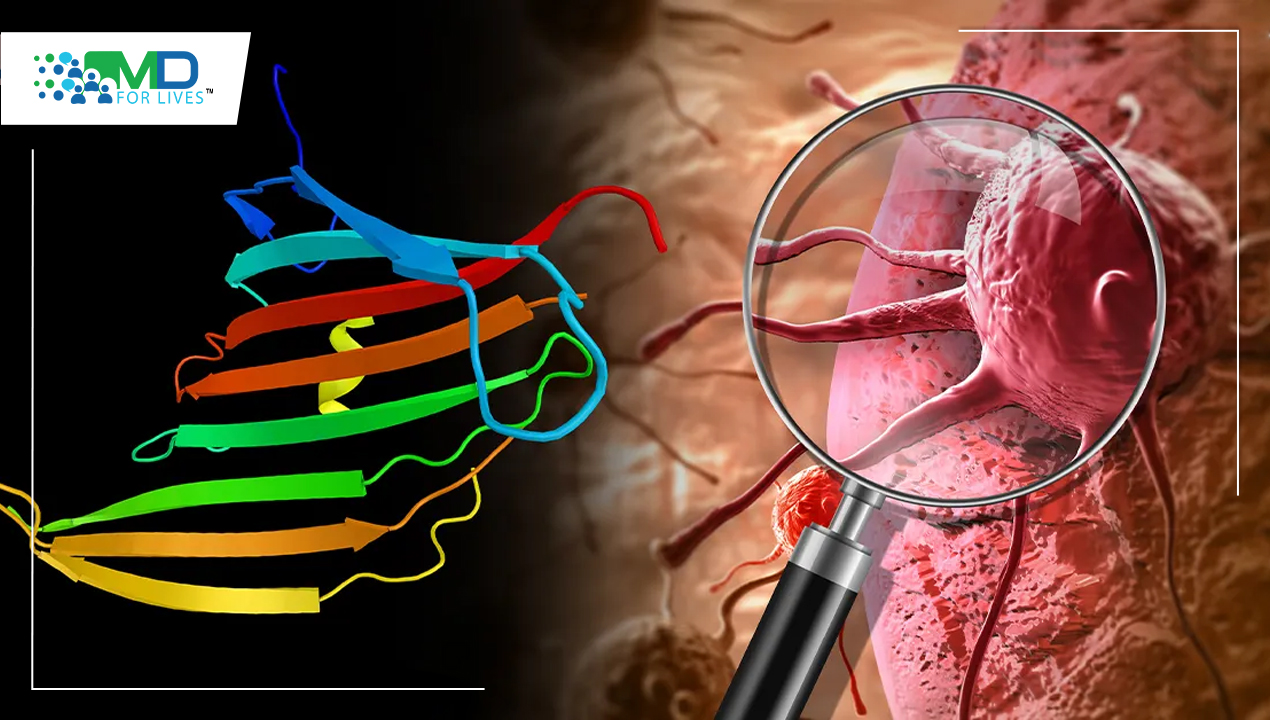
Our cells are protected from cancer by the p53 protein, which is a promising target for cancer therapy. The concern is that it degrades quickly inside the cell. Researchers at Sweden’s Karolinska Institutet have discovered a unique method of stabilizing the protein and increasing its potency. They demonstrate that by combining p53 with a spider silk protein, they may develop a protein that is more stable and capable of killing cancer cells.
Cancer Suppressing Protein p53
TP53 is a gene that produces a protein that is present inside the nucleus of cells and is involved in cell division and death control. The P53 gene directs the production of a tumor protein p53 (or p53). This protein has anti-tumor properties. p53 is a pro-apoptotic transcription factor found in the nucleus. Because the p53 gene exhibits loss of function mutations in approximately 50% of human malignancies, it is considered one of the classical tumor suppressors. Wild-type p53 is inhibited by mutant p53, which functions as a dominant-negative inhibitor. Mutant p53 does, in fact, have oncogenic potential. Chemo-resistant phenotype is seen in some malignant cancer cells with p53 mutations.
P53 is stimulated to accumulate in the cell nucleus in response to a range of cellular stressors, such as DNA damage, to perform its pro-apoptotic role. Through the transactivation of its target genes implicated in the induction of cell cycle arrest and/or apoptosis, activated p53 promotes cell cycle arrest to allow DNA repair and/or apoptosis to limit the propagation of cells with substantial DNA damage. As a result, p53’s DNA-binding activity is intimately connected to its tumor-suppressing function.
Spider Silk Protein
For thousands of years, spider silk has piqued human attention, owing to its tenacity and ductility, as well as the fact that it does not appear to produce irritation or allergic reactions. As a result, spider silk has been used for bandages as well as hunting and fishing.
Spiders employ their silk for a variety of purposes in nature, including webs, wrapping prey, protecting their offspring, and as a lifeline to enable their safe escape from predators. Spiders have a significantly wider range of silk usage than insects like silkworms, which primarily use their silk to make cocoons.
Spider silk is mostly made up of proteins with a lot of nonpolar and hydrophobic amino acids like glycine and alanine, but no or very little tryptophan, for example.
Short polypeptide segments of roughly 10–50 amino acids make up the repeated sequences, which account for more than 90% of the total spider silk protein. Within a single protein, these motifs can be found more than a hundred times. As a result, each polypeptide repeat has individual functional characteristics, resulting in spider silk strands with exceptional mechanical qualities.
Spidrons—floppy proteins spun into silk by spiders—have been studied by scientists in the hopes of developing future cancer treatments.
Read about Amino Acid Sensor
Fusion of Spider Silk Protein and p53 can Prevent Cancer
P53, a protein that plays an important role in the body’s immunological response to cancer, is a promising target for cancer treatment. Specifically, our systems rely on p53 to keep cancer cells from rapidly expanding and dividing.
Intrinsically disordered domains (IDDs) are proteins that do not assume a well-defined three-dimensional structure and are engaged in a range of important cellular activities, yet they are disproportionately abundant among cancer-related proteins.
IDDs’ propensity to participate in a wide range of interactions, from transcription factor recruitment to cell signaling, makes them appealing therapeutic targets, but it hampers structural biology and drug design attempts. Perhaps the most prominent example is the tumor suppressor p53, which plays a central role in the control of cell proliferation. Because inactivating p53 mutations are seen in up to 60% of all human malignancies, restoring its activity is an appealing therapeutic option. Its N-terminal region is chaotic by nature.
In human cells, N-terminal abnormality is a negative regulator of protein half-life and is a characteristic of proteins that are vulnerable to fast turnover, which is consistent with p53’s short life cycle in healthy cells. As a result, p53 expression levels in human cells are low at baseline, implying that there is no evolutionary drive to adopt a stable shape. As a result, p53 has a high proclivity for aggregation in vitro and can spontaneously assemble into amyloid-like fibrils. As a result, it is an excellent example of a protein that is “on the verge of solubility.”
Stabilizing cellular p53 is a viable cancer treatment strategy. The proteins that make up spider dragline silk, known as major ampullate spidroins, are produced in the silk gland and stored in a soluble form in the sac at concentrations surpassing 40% w/v. They have a 130-residue N-terminal domain (NT) that is highly conserved, followed by several hundred poly-alanine and glycine-rich stretches and a regulatory C-terminal region. Through pH-dependent dimerization, the tightly folded, α-helical NT promotes spidroin assembly. We recently showed that a non-dimerizing mutant increases expression while suppressing self-assembly, allowing the efficient generation of aggregation-prone peptides.
The spider silk domain increases p53 translation by causing its intrinsically disordered N-terminal domain to assume a compact shape. Spider proteins, which are spun into silk by spiders, are similar to p53. They are also big and floppy, and they cluster together easily. They are, however, unlike p53, capped by a short, compact component (called a domain) that is very stable and can be easily produced by the cellular protein manufacturing machinery.
The human p53 protein had a short portion of a spider silk protein – a domain – linked to it. When this “fusion protein” was given to cells in the lab, it was discovered that they produced vast amounts of it. The most floppy region of the p53 protein was discovered to be wrapped around the spider silk domain like a thread around a spindle. By “wrapping up” the protein in this way, the spider silk domain was able to pull it out of the cellular production machinery, resulting in the creation of more protein.
To see if the spider silk-p53 protein is functioning, researchers placed it into cancer cells with “reporter genes,” which force the cell to light up when p53 activates genes that allow the cell to stop growing. Surprisingly, the fusion protein elicited a higher response than conventional p53, implying that the spider silk domain may theoretically be exploited to boost p53’s ability to shut off cancer cells. If the RNA, which contains the genetic “blueprint” for making p53, is given to cells, it may include modified spider silk domains that boost the cells’ ability to produce the protein.
The method is intended to be adapted in the future to target a wide spectrum of partially disordered proteins, allowing structural research of particularly difficult systems.